Speeding Up Motif Finding
This problem asks:
Given: A DNA string s.
Return: The failure array of s.
Required reading
Restate the problem
The Knuth-Morris-Pratt algorithm is an advancement in the efficiency of finding a substring within a given string. The central improvement over considering each individual character in the string as a possible beginning of the substring is that the Knuth-Morris-Pratt algorithm keeps track of partial successful matches. If a section of the string matches the substring, but then fails to match midway through, the Knuth-Morris-Pratt algorithm doesn’t start over with the next character in the string. Instead, it skips ahead to the first character that could possibly be a start of the substring.
For this challenge, I’m going to get a DNA string, s. I need to return a failure array of integers the same size as s. Every number in the failure array is the count of characters at that point in the string s that match the beginning of the string s.
The sample dataset and sample output are helpful in understanding what’s being asked here.
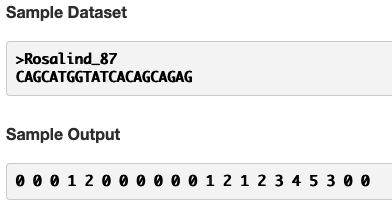
The first value in a failure array is always 0. We’re given that value in the problem statement. The second value is a 0 because A, the second character, does not match C, the first character.
The third value is 0 because G, the third character, does not match C, the first character.
The fourth value is 1 because C, the fourth character, matches C, the first character.
The fifth value is 2 because A, the fifth character, matches A, the second character.
The sixth value is 0 because T, the sixth character, does not match G, the third character.
… and so on.
Solution steps
First, I read in the DNA string and create a result array that’s the same length as the DNA string with a 0 in every position.
Then I start with the second value in the string and increment that value by 1 if it matches the start of the string.
Then I continue iterating through the string and incrementing the positions in the failure array every time there is a match between the current character and the next character from the beginning of the string.
At the end, I throw out the first value in the array and add a 0 at the end. Why?
Well, that’s a bug.
I threw out the first value and added a 0 at the end when I was working on this a few weeks ago because that’s what I needed to do to make my code return the correct result for the sample dataset. When I ran it on the real dataset and got a correct response, I stopped working on this and moved on. Now that I’m looking at it more closely to write this article, I see a lot about my solution that I don’t like. In fact, I think I was lucky to get a dataset that ended with a failure array of 0, because otherwise, my code won’t work.
Am I going to dig into code that returned a correct result and make sweeping changes just because I want to make sure I have reliable code to a challenge that I’ve already solved?
Yes. Yes, I am.
For reference, the sketchy code that returned a correct result is here.
I’ll keep track of my actions here: I saved a copy of the correct response to my first downloaded dataset so that I can check to see if my altered code still returns a correct result.
I simplified reading the downloaded DNA string from this:
sequence_collection = []
for seq_record in SeqIO.parse(file_path, "fasta"):
sequence_collection.append(seq_record.seq)
sequence = sequence_collection.pop(0)
to this…
for record in SeqIO.parse(file_path, "fasta"):
sequence = record.seq
I got rid of this line without replacement because throwing away the first value in the result might make sense, but there’s no reason to add a zero onto the end.
solution = str((result[1:], 0))
Instead of starting with position = 2 and using a while statement, I changed to starting with position -1 and using a for-while statement. This required a complete rewrite. The old code:
result = [0]*len(sequence)
position = 2
condition = 0
while position < len(sequence):
if sequence[position-1]==sequence[condition]:
condition+=1
result[position] = condition
position+=1
elif condition>0:
condition=result[condition]
else:
result[position]=0
position+=1
changed to the new code:
result = [-1]
counter = -1
for i in range(len(sequence)):
while counter >= 0 and sequence[i] != sequence[counter]:
counter = result[counter]
counter += 1
result.append(counter)
print(*result[1:])
I replaced this line:
print(str(result).replace('(', '').replace("[",'').replace(',','').replace(']','').replace(')',''))
with this:
print(*result[1:])
I needed to get rid of the first value in the result list because it was the -1 that I put there before processing as a placeholder for the counter.
I verified that the new code returned correct responses for both the sample dataset and the real dataset from my original solution. Then, just to be extra-sure I had everything right, I downloaded another real problem dataset from Project Rosalind and submitted my correct result from the new code.
Post-solution notes
Challenges solved so far: 45
How many people solved this before me: 2,833
Most recent solve before me: 3.5 hours
Time spent on challenge: 2 hours, mostly on overhauling the code while writing the article
Most time-consuming facet: rewriting the code after solving
Accomplishments and badges: String algorithms badge level 3, shown below
Closing thoughts: I’m glad I rewrote my code even though it worked once before. I feel much better about this because I understand how it works.