Calculating Protein Mass
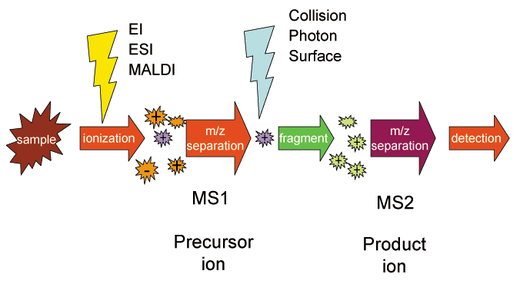
image of tandem mass spectrometry from Wikipedia
This problem asks:
Given: A protein string P of length at most 1000 aa.
Return: The total weight of P.
Required reading
Restate the problem
There is a monoisotopic mass table provided with the challenge. For each protein in the string of proteins they provide, I need to look up the weight in the table and add all the weights together.
Solution steps
I decided to use Biopython instead of writing my own table look-up code, so the first step was to find the right Biopython method to use.
The Bio.SeqUtils.ProtParam module includes a molecular_weight method which is an exact fit for this challenge.
When I set the isotope mode to True and subtracted 18.01056 da from the final result to account for the single water molecule mentioned in the challenge, I got the correct weight for the sample dataset.
Python concepts
My solution code for this challenge is here.
There were no new Python concepts used in this solution.
Writing the look-up code in Python would have used a dictionary to hold the molecular mass table, then an algorithm like this to look up the masses and add them together:
mass = 0
for char in protein:
mass += amino_mass[char]
Bioinformatics concepts
Although adding these protein masses together, discovering the weights of the different proteins took a lot of work.