Counting DNA Nucleotides
This problem asks:
Given: A DNA string s of length at most 1000 nt.
Return: Four integers (separated by spaces) counting the respective number of times that the symbols ‘A’, ‘C’, ‘G’, and ‘T’ occur in s.
Restate the problem
They’re going to send me a DNA string no longer than 1000 characters. I need to count the A’s, C’s, G’s, and T’s, then return those results.
Solution steps
Python includes a list count method that solves this problem in one step.
I used python’s list count method to count all the A’s, C’s, G’s, and T’s as shown below.
return (
sequence.count("A"),
sequence.count("C"),
sequence.count("G"),
sequence.count("T")
)
Then I wrote my results to a text file: problem solved.
Python concepts
Efficiency My solution ran on the test set in less than a second, but it’s not the fastest solution because it has to go all the way through the test set four times. Once to count the A’s, once to count the C’s, etc…
Since the problem tells us the maximum number of letters in the test set is 1000, and we have 5 minutes to solve the problem, my inefficient solution that takes less than a second is fine.
If I wanted to solve this problem for datasets that are many orders of magnitude larger, I would set up four counters: A, C, G, and T. Then I would run through the list one time and increment the counter that matches the letter in the test set.
File handling Python comes with easy-to-use tools for opening, reading, appending, writing, and closing files. Using the file handling functions was straightforward. I followed the examples in the documentation, and they worked as expected.
Idempotency
Wikipedia article on Idempotence
Idempotent has many different meanings. The one that applies here is that once an idempotent process has been executed once, executing it again has no effect.
One day-to-day example of an idempotent function is an elevator call button. Pressing it the first time calls the elevator. Pressing it any number of times after that has no effect.
It’s useful for computer programs to be idempotent because it means the operation can be tried as often as necessary without unintended effects.
To make my program idempotent, I check to see if there’s already a solution file in place before I try to write a new solution file.
if os.path.exists(solution_path):
os.remove(solution_path)
If I don’t check, and there’s already a solution file there, I’ll get an error:
Traceback (most recent call last):
File "/Users/robertbryan/PycharmProjects/rosalind/solution-code/dna.py", line 29, in <module>
file = open(solution_path, "x")
FileExistsError: [Errno 17] File exists: '../solution-outputs/rosalind_dna.txt'
There are several ways to avoid this error.
Bioinformatics concepts
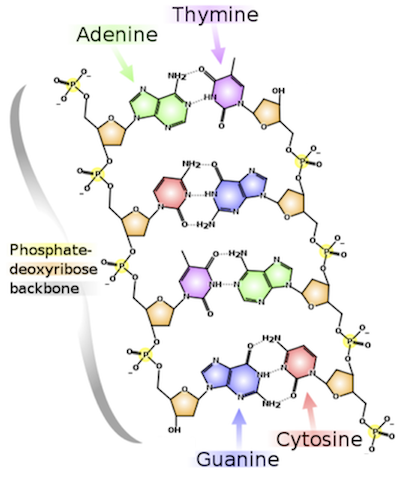 image from Project Rosalind
image from Project Rosalind
While counting the different letters in a string of text is relatively simple, DNA structure that those letters represent is fascinating and complex. This problem introduces the concept of the DNA code with carries genetic information for the transmission of inherited traits.